NPM1 mutations are acquired driver mutations that play a critical role in the pathogenesis of NPM1m AML.2,3 In this distinct genetic subtype of AML, menin interacts with wild-type KMT2A (MLL1) and can contribute to leukemogenesis.1,4,5
NPM1 mutations are the most common genetic alteration in adult AML cases, and prognosis after relapse is poor.3,4
NPM1 mutations are found in ~35% of patients with AML3,6
Relapsed/refractory NPM1m AML is an acute leukemia with a critical unmet need
Current guidelines recommend testing for NPM1 mutations as part of the diagnostic workup of acute leukemia6
Therapeutic interventions are urgently needed as there are currently no approved treatments that selectively target the underlying disease mechanisms driving NPM1m AML2,4
NPM1m AML has a strong HOX/MEIS1 gene expression pattern with a dependency on the menin-KMT2A interaction.2
In NPM1m AML, nuclear NPM1m binds to chromatin and abnormally exposes transcription start sites for menin-KMT2A binding, leading to upregulation of HOX/MEIS1 gene expression, resulting in leukemogenesis.7
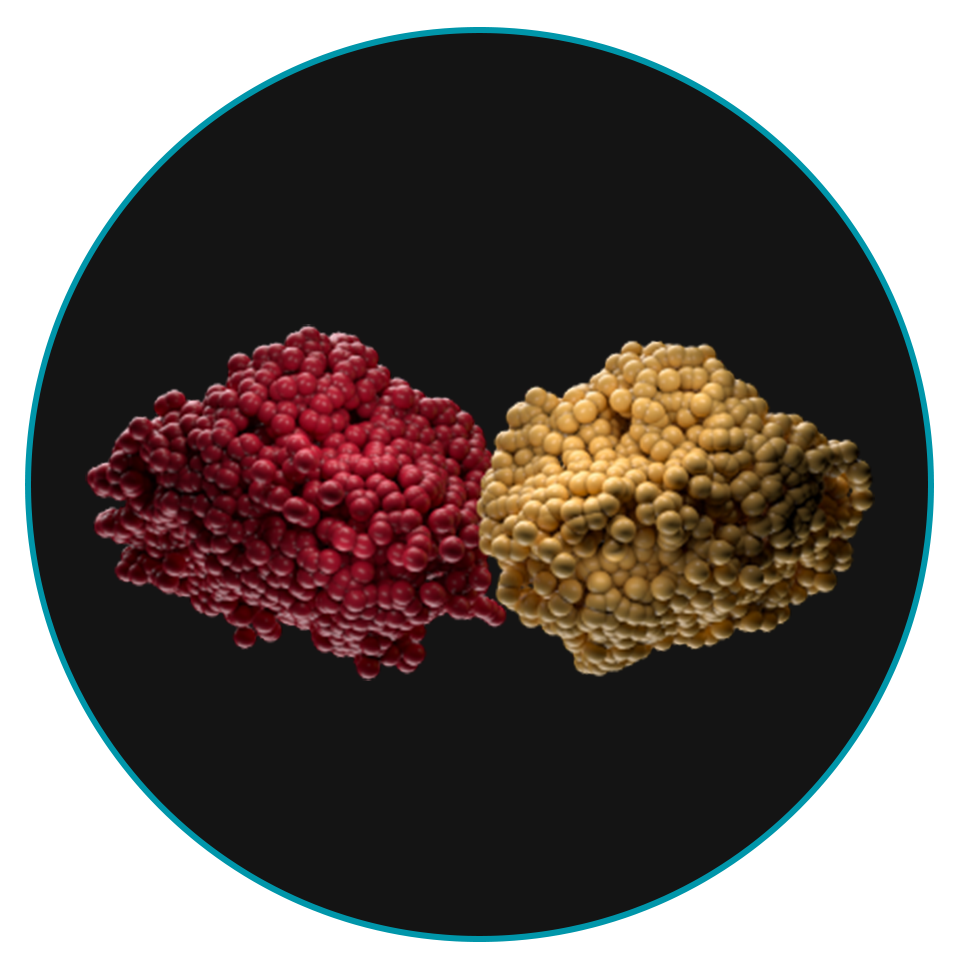
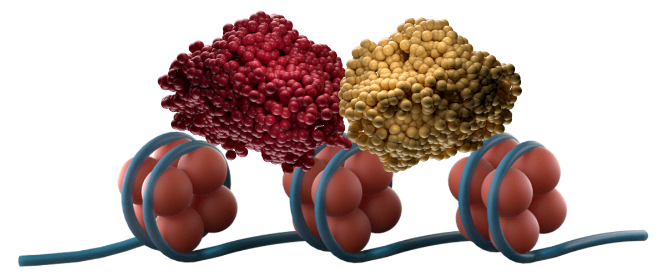
Menin interactions modify chromatin, allowing the menin-KMT2A complex to drive increased expression of HOX and MEIS1 genes7

Upregulation of HOX/MEIS1 gene expression promotes leukemogenesis7

Increased proliferation of undifferentiated cells results in acute leukemia7

 In NPM1m AML, menin interacts with
In NPM1m AML, menin interacts with
Menin interactions modify chromatin, allowing the menin-KMT2A complex to drive increased expression of HOX and MEIS1 genes7
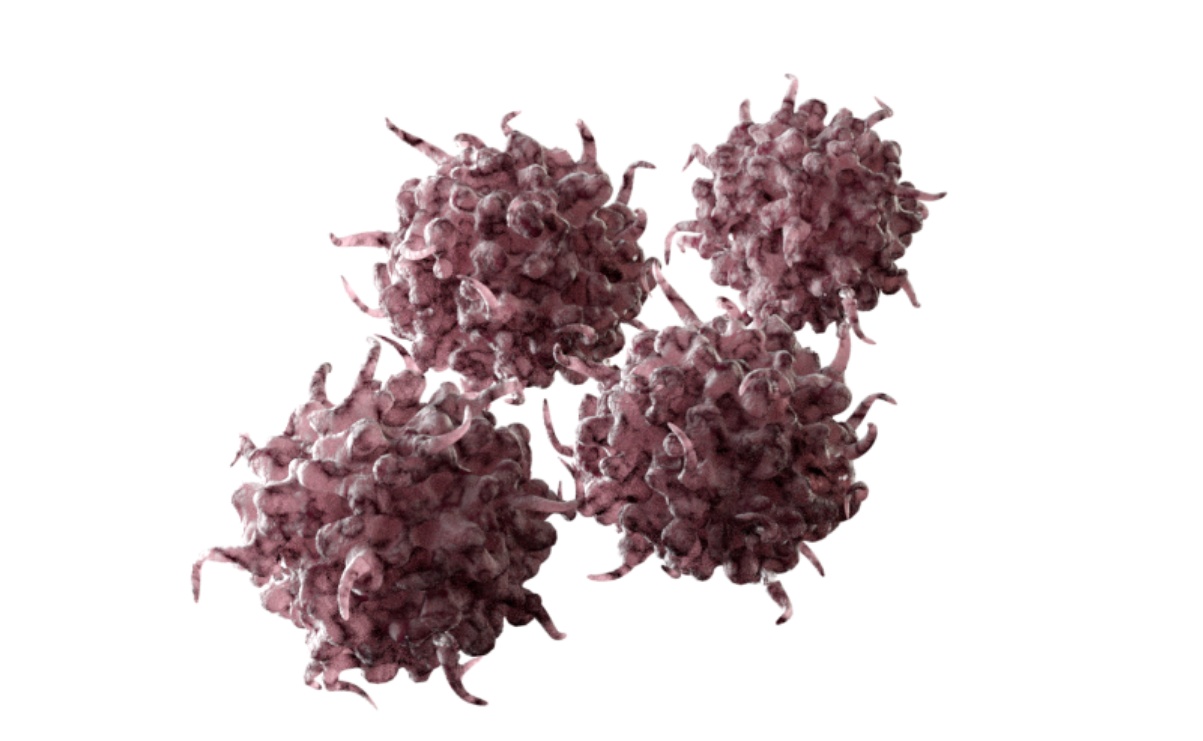
Upregulation of HOX/MEIS1 gene expression promotes leukemogenesis7

Increased proliferation of undifferentiated cells results in acute leukemia7
With a deeper understanding of the mechanisms that contribute to NPM1m AML, the menin-KMT2A interaction becomes an intriguing target to investigate further.
AML=acute myeloid leukemia; HOX=homeobox; KMT2A=lysine methyltransferase 2A; MEIS1=MEIS homeobox-1; MLL1=mixed-lineage leukemia protein-1; NPM1m=mutant nucleophosmin-1.
References: 1. Issa GC, et al. Leukemia. 2021;35:2482-2495. 2. Falini B. Am J Hematol. 2023;98(9):1452-1464. 3. Ranieri R, et al. Leukemia. 2022;36(10):2351-2367. 4. Kühn MWM, et al. Cancer Discov. 2016;6:1166-1181. 5. Uckelmann HJ, et al. Science. 2020;367:586-590. 6. Arber DA, et al. Arch Pathol Lab Med. 2017;141:1342-1393. 7. Uckelmann HJ, et al. Cancer Discov. 2023;13(3):746-765. 8. Yokoyama A, et al. Cell. 2005;123:207-218.